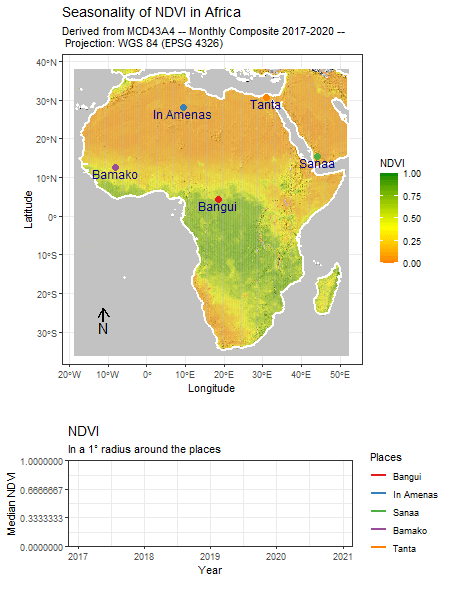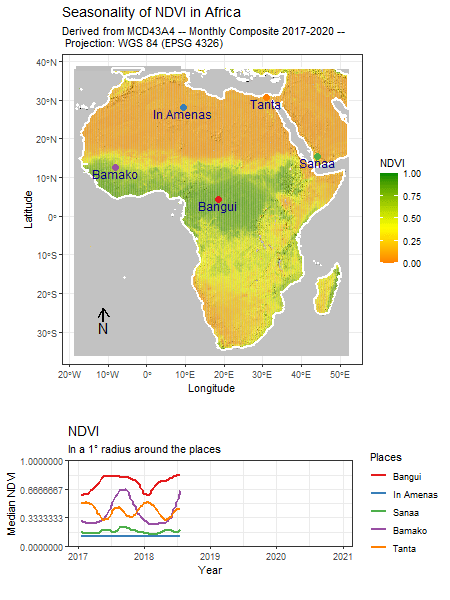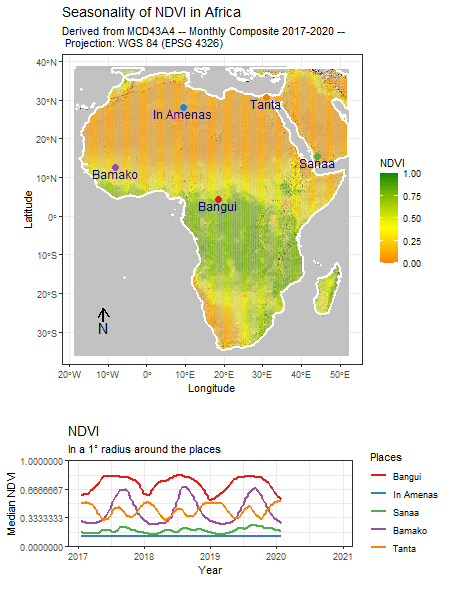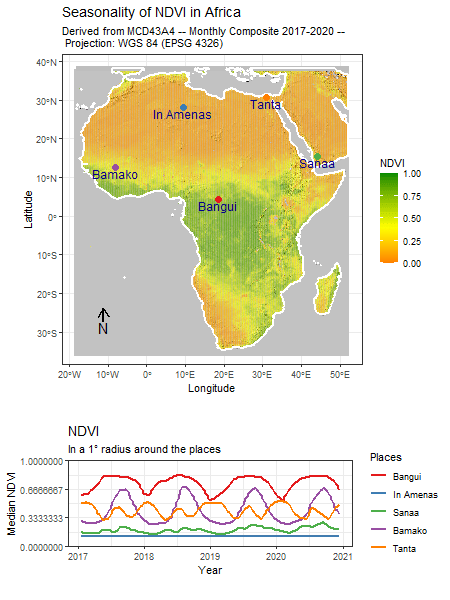
Category: R
Book “Intro to Spatial Data Analysis” ...
Posted by Martin Wegmann | Jun 10, 2020 | Biodiversity, Conservation, News, publication, R, Software, Students | 0 |
Upcoming book “Introduction to Spatial Data ...
Posted by Martin Wegmann | Mar 4, 2020 | Biodiversity, Conservation, News, publication, R, training | 0 |
PhD position: Earth Observation of drought and fire impacts
by EORhub | Dec 19, 2024 | Africa, Biodiversity, Conservation, PhD, position, R, UAV | 0 |
Job Announcement: PhD Position on EO research of Drought, Fire and Vegetation in Kruger National...
Read MoreCourse on Advanced Remote Sensing taught at FUT Minna
by Michael Thiel | Jul 26, 2020 | events, News, project, R, training, WASCAL-DE-Coop, West Africa | 0 |
Our colleagues Steven Hill and Dr. Michael Thiel were giving a two weeks online course on Advanced...
Read MoreLecture on Advanced remote Sensing in Ghana
by Michael Thiel | Jun 20, 2020 | events, News, project, R, training, WASCAL-DE-Coop, West Africa | 0 |
Our staff members Dr. Frank Thonfeld, Steven Hill, and Dr. Michael Thiel taught a three-week...
Read MoreBook “Intro to Spatial Data Analysis” in Print
by Martin Wegmann | Jun 10, 2020 | Biodiversity, Conservation, News, publication, R, Software, Students | 0 |
Our upcoming book “Introduction to Spatial Data Analysis” is finally in print and...
Read MoreVisualizing movement trajectories in R using moveVis: Article published in the latest issue of Methods in Ecology and Evolution
by Jakob Schwalb-Willmann | May 12, 2020 | News, publication, R, Software | 0 |
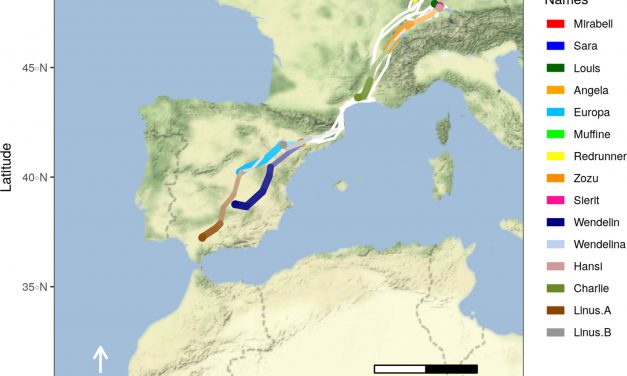
Figure 1: Migratory movements of white storks Ciconia ciconia on a Mapbox satellite base map...
Read MoreUpcoming book “Introduction to Spatial Data Analysis” – 30% discount for pre-order
by Martin Wegmann | Mar 4, 2020 | Biodiversity, Conservation, News, publication, R, training | 0 |
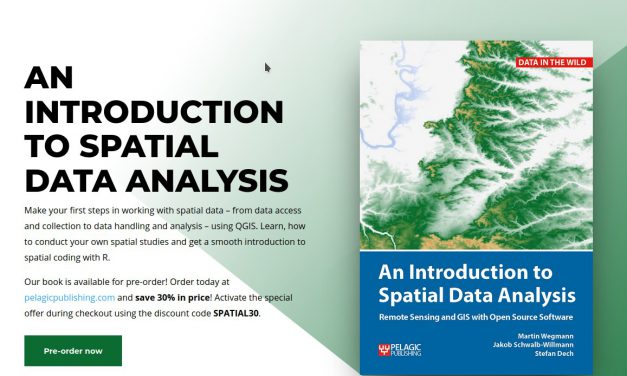
Our book is available for pre-order! Order today at pelagicpublishing.com and save 30% in price!...
Read MoreDevLab 2020: EO Data Cube Workshop by Steven Hill
In our Department’s first DevLab workshop of 2020, Steven Hill will give an introduction into the Open Data Cube infrastructure of our Department and demonstrate the cube’s web and Python user interfaces.
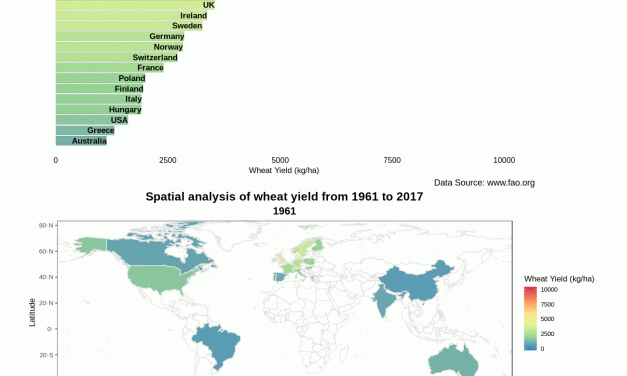
Read MoreBar Chart race using ‘gganimate’ for world’s most efficient wheat yield producing countries
by Maninder Singh Dhillon | Apr 21, 2019 | Agriculture, News, R | 0 |
Easy spatio-temporal visualization of 15 selected efficient wheat yield producing countries using...
Read More
news blog by
the Earth Observation Research cluster
EORC
at the University of Würzburg
Institute of Geography and Geology
John Skilton Str 4a
97074 Würzburg
recent posts
-

-

-
 The “Geolingual Studies” team visited the DLR EOCJul 6, 2025 | project
The “Geolingual Studies” team visited the DLR EOCJul 6, 2025 | project -
 New PhD student Tamilwai KolowaJul 5, 2025 | staff
New PhD student Tamilwai KolowaJul 5, 2025 | staff -
 Course on Object-based image analysisJul 4, 2025 | News
Course on Object-based image analysisJul 4, 2025 | News